Artificial intelligence (AI)/machine learning (ML) methods can now predict the 3D folded structures of many proteins with surprising accuracy, potentially shortening the time from inspired ideas to actual medical treatments. What used to be a long and uncertain path to obtain the structures of targets for drug discovery can now be reduced significantly. These advances in understanding protein folding unlock new opportunities to accelerate drug discovery, especially when combined with Schrödinger’s physics-based computational platform.
Beyond the blueprint
In the time since scientists finished mapping the human genome, we’ve learned that merely having the DNA blueprint isn’t enough. To design drugs, we need to understand proteins, which are the molecular machines that underlie the many functions of each of our cells. What are the physical principles governing the 3D shapes of proteins, and how do their structures control cellular functions? How do drugs interact with proteins in a way that therapeutically benefits the body? The new frontier of discovery is now with the scientists—including those at Schrödinger—who are pioneering the deepest understanding and largest exploration of protein folding, form and function to date.
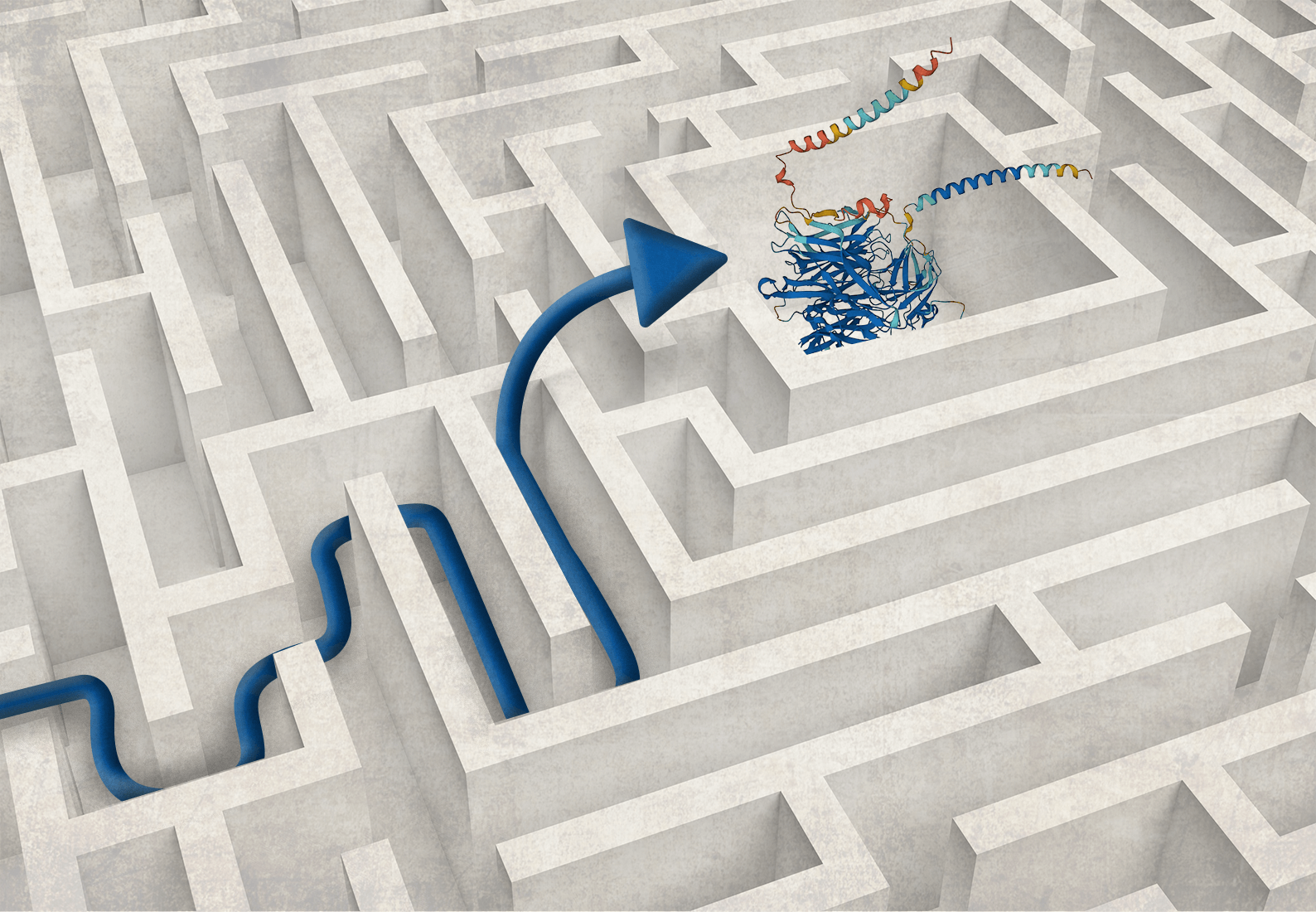
The long slog: Tireless explorers in a massive labyrinth
In the past, scientists didn’t have the luxury of access to massive amounts of information about the 3D structures of proteins. Structural biologists have cumulatively conducted decades of experimental work using techniques such as X-ray crystallography and cryo-electron microscopy to give us a wealth of knowledge that continues to grow each day. This explosion of data is stored in a massive public data set, the Protein Data Bank (PDB), a repository that needed to be tamed in order to be properly mined.
Artificial intelligence (AI) company DeepMind accepted the challenge. AI is a broad term referring to computers completing tasks normally performed by humans. The term machine learning (ML) is often used interchangeably with AI, but machine learning is the more specific branch of AI involved in drug discovery. Machine learning systems are designed to comb through large amounts of existing data, enabling computers to detect patterns, predict outcomes, and make decisions without being specifically programmed.
AI is a broad term referring to computers completing tasks normally performed by humans. The term machine learning (ML) is often used interchangeably with AI, but machine learning is the more specific branch of AI involved in drug discovery.
DeepMind researchers have taken the wealth of public structural data from the PDB, plus amino acid sequences, and developed a new machine learning language that can learn millions of parameters directly by training networks that generate 3D protein structures. Called AlphaFold, this tool helps other scientists by giving them access to detailed models of protein structures that were previously unavailable. At the same time, an academic group of scientists at the University of Washington created a tool called RoseTTAFold, which offers similar capabilities. More recently, researchers at Meta (formerly Facebook) reported ESMFold, which promises to be faster than AlphaFold and RoseTTAFold while delivering similarly accurate models. There are now also community driven efforts for similar open source protein structure prediction tools, like OpenFold, which provides additional training pipelines for fine tuning protein folding structure models using sequence databases.
Although researchers still don’t always have atomic-level details of certain proteins that are targets for new drugs, the new insights from these AI/ML-based models are a significant step forward. These tools now give drug hunters and experimentalists not only good starting points to begin a drug discovery project, but also the ability to do better, smarter, and optimized experiments that will lead to more efficient discoveries.
While the most expensive part of new-drug creation remains human trials, the contributions of AI/ML-based protein-prediction software can help drugmakers get to the testing stage faster. These AI/ML-based methods make the journey more efficient by letting researchers start with the most promising paths for target identification. In a way, scientists can now “peek” down each path and decide if they want to proceed or if they want to keep looking for other paths.
What’s so important about how proteins are folded?
Proteins are the business end of the genome. They’re the driving force for bodily functions. We know that proteins are made from amino acids joined together and are differentiated by how those building blocks are organized, ordered, and connected. The machine-like, action-based nature of proteins comes from the arrangement in space of atoms making peptide bonds, side chains, and the physico-chemical attributes of the resulting 3D shape.
Researchers who evaluate the minuscule 3D structures of proteins can learn the most when the protein is in sharp focus. That way, the structure—or the position of each atom in the protein—is known with very high accuracy, which is an essential starting point to design potent and selective drugs. Structural biology has been instrumental in the discovery of many approved drugs, and more and better structures are constantly needed in order to make new and better medicines for unmet medical needs.
Schrödinger’s physics-based modeling software can then be used to extract the most value and information out of both experimental and AI/ML-predicted structures.
AI/ML and physics-based modeling
To effectively design drugs that target a protein, chemists need high resolution protein structures (below ~3 Å, with higher-precision atomic positions corresponding to a resolution range of ~1-2 Å). Even if AI/ML-based methods can now provide protein structure models with unprecedented accuracy, they are still far from perfect. In a recent publication, scientists from the Protein Data Bank estimated that the virtual structures of AlphaFold and RoseTTAFold predictions are as accurate as ~3.5 Å resolution experimental structures. And that’s where synergies between machine learning, structural biology, and physics-based modeling can help.
The availability of these accurate theoretical protein models can aid and speed up the experimental determination of structures of the drug molecule candidates bound to the protein target. This is an essential step for a drug discovery project, and it is a current limitation of AI/ML-based protein-prediction tools.
Schrödinger’s physics-based modeling software can then be used to extract the most value and information out of both experimental and AI/ML-predicted structures. These are static representations, and they lack many key physico-chemical details of the local environment and dynamic properties of proteins that are important to decipher and target their functions. By taking these into account, molecular modeling methods that are based on physics provide a more realistic picture of protein folding and proteins’ behavior.
The combination of AI/ML and physics-based modeling technologies means fewer dead-end leads and far more active ones. Using these together will provide a better picture of protein folding and how proteins will likely work in real-world conditions, enabling more effective design of drugs that can interfere with protein malfunction and treat disease, ultimately facilitating drug discovery and reducing risks along the way.