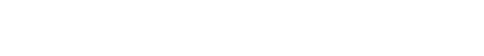Like many things in life, early-stage drug discovery is a numbers game. The more chemical space that can be explored earlier in the discovery process, the greater the likelihood of finding novel, biologically active molecules that meet key parameters for clinical advancement. Historically, this exploration involved screening just thousands to a few million compounds through costly, automated experimental testing. To put this into perspective, chemists estimate there are 1060 possible drug-like compounds that could be synthesized – this is double the number of stars estimated in the universe. Using classic screening approaches, exploring this scale of chemical space is fundamentally cost prohibitive and technically impractical.
To overcome this challenge, scientists are increasingly turning to alternative methods that can screen billion to trillion compound libraries using new experimental techniques such as DNA-encoded libraries (DELs) and new computational techniques such as machine-learning enhanced virtual screening. Multiple studies using these methods have since proven what appears to be intuitively obvious: screening significantly more compounds yields a greater diversity of novel hits.1 More shots on goal increases the chances of success.
The popularity of DNA-encoded libraries have grown dramatically over the last several years as the method allows screening of tens of billions (potentially up to trillions) of molecules in a single tube and produces massive quantities of valuable quantitative data. The challenge with DEL screening, however, is that the results can be plagued by false negatives and noisy data, which can lead researchers to overlook the most promising candidate molecules. This noise-level increases linearly with the size of the DEL library – larger screens are noisier than smaller ones. Additionally, DEL screens often result in highly ranking molecules that have a poor mix of drug-like properties and can prove challenging for medicinal chemistry intuition during hit selection. Together, these challenges can hamper the success of some DEL screening efforts.
Last year, Schrödinger teamed up with WuXi AppTec, a leading provider of DEL screening solutions, to mitigate these challenges and develop methods to identify only the most promising drug-like compounds by complementing DEL screens with machine learning and computational workflows. Starting with results from a DEL screen of 15 billion compounds, the teams posed the questions: Can we select better compounds using machine learning-enhanced methods, and can we increase the pool of candidates with better physicochemical properties?
Using a series of advanced machine learning methods to train against the vast amount of bioactivity data produced from the DEL screen, the team built a model of compound potency with the goal to amplify the signal of active hits in the initial experimental results. Additionally, the team performed a workflow to filter based on drug-like properties using physics-based methods.
The study results show that by complementing DEL screens with machine learning workflows, Schrödinger scientists were successfully able to:
- Prioritize compounds using a machine learning potency model and recover more missed hits (reduce false negatives
- Optimize physiochemical and ADME properties while balancing potency to identify compounds with the most promising drug-like characteristics
- Identify additional potent compounds within the same chemical space as the DEL screen using generative methods to ideate novel putative active compounds.
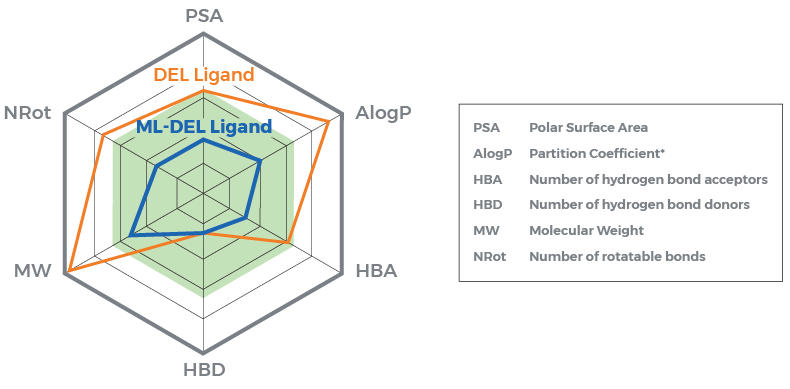
Figure: Results showed improved combination of chemical properties from the top scoring machine learning-enhanced DEL ligand compared to original top scoring molecule from DEL screen.
The results highlight the complementarity of high throughput screening and machine learning approaches to maximize the discovery of novel hits with the most promising combination of properties. Based on the success of this method, Schrödinger has partnered with WuXi to make these machine learning-enhanced workflows available via research services from Schrödinger for immediate use with DELlight screens.
_______